Decoding the evolution and genomics of sleep
Using comparative approaches to understand the evolution of sleep
The biological mechanisms that regulate sleep are conserved across vertebrates, yet sleep displays remarkable variation both across and within species. Why do animals display variation in sleep, and what genes and cell types drive such variation?
We study the evolution and genomics of sleep variation across fish animals using a two-pronged macro- and micro-evolutionary approach. We leverage the amazing diversity in sleep related phenotypes that we have discovered across both African cichlid fish (micro-evolution), and across the larger radiations of bony and cartilaginous fishes (macro-evolution).
We have a commitment to fostering a comfortable and safe environment for people to perform their best. As a group, we believe in continually educating ourselves and striving towards a more inclusive scientific culture.
Research Overview
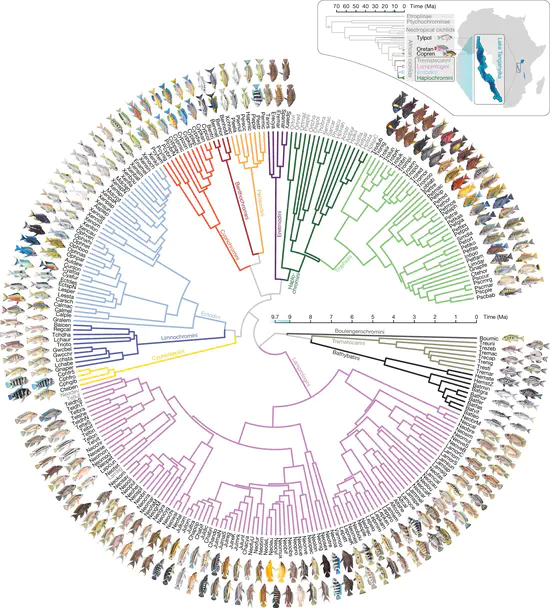
Micro-evolutionary comparisons across Lake Tanganyika cichlid fishes
Cichlids represent the most extraordinary example of a vertebrate explosive adaptive radiation — approximately 2000 species have evolved within the last 10 million years in the Rift Valley Lakes of Africa. We have recently demonstrated that these cichlids display incredible diversity in sleep-related behaviours (sleep timing, sleep duration). Work in this area of the lab is focused on understanding the evolution, ecologies, and molecular mechanisms underlying this varition.

Macro-evolutionary comparisons across fish and vertebrates
What is the evolutionary history of sleep timing in fish and other vertebrates? What genomic, ecological, or paleontological events are associated with shifts in sleep timing in over >400my of vertebrate evolution?
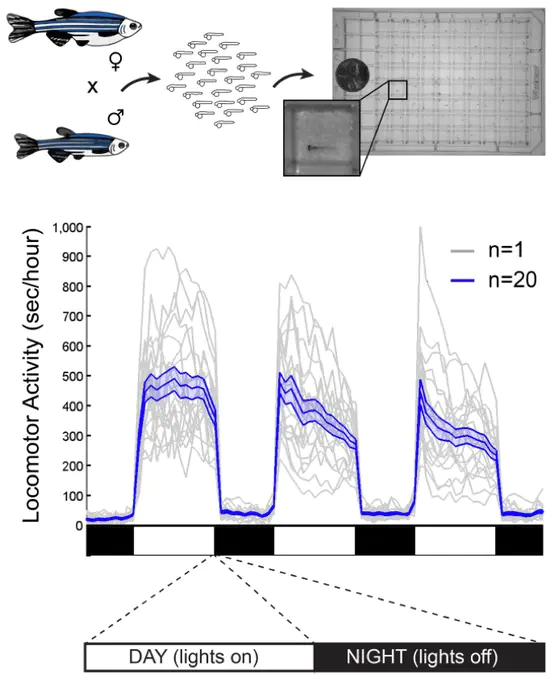
Identifying and characterising new sleep genes
We use aim to use a combination of traditional model organisms, as well as functional genomic, behavioural, molecular, and circuit based techniques to characterise novel sleep genes and circuits identified through our evolutionary comparisons.
Selected publications
*Corresponding author, †Contributed equally, Shafer Lab members
- Nichols, A.L.A.†, Shafer, M.E.R.†*, Indermaur, A., Rüegg, A., Gonzalez-Dominguez, R., Malinsky, M., Sommer-Trembo, C., Fritschi, L., Schier, A.F., Salzburger, W. Widespread temporal niche partitioning in an adaptive radiation of cichlid fishes. Nature Ecology & Evolution (in press). 2025. DOI
- Shafer, M.E.R.*, Nichols, A.L.A, Schier, A.F., Salzburger, W. Frequent transitions from night-to-day activity after mass extinctions. bioRxiv. 2023. DOI
- Do nocturnal habits help protect animals from extinction?
- Burguera, D., Dionigi, F., Kverková, K., Winkler, S., Brown, T., Pippel M., Zhang, Y., Shafer, M.E.R., Nichols, A.L.A, Myers, E., Němec, P., Musilova, Z*. Expanded olfactory system in fishes capable of terrestrial exploration. BMC Biology. 2023. DOI
- Shafer, M.E.R.*, Sawh, A.N., Schier, A.F.* Gene family evolution underlies cell type diversification in the hypothalamus of teleosts. Nature Ecology & Evolution. 2021. DOI
- Sawh, A.N.*, Shafer, M.E.R., Su J.H., Wang, S., Mango, S.E.* Lamina-dependent stretching and unconventional chromosome compartments in early C. elegans embryos. Molecular Cell. 2020. DOI
- Shafer, M.E.R.* Cross-species analysis of single-cell transcriptomic data. Frontiers in cell and developmental biology. 2019. DOI
Interested in joining the lab?
If you are interested in contributing to, working with, or joining the Sleep Evolution Group in any capacity please reach out to Dr. Maxwell (Max) Shafer. In general, my training philosophy is to provide each trainee with opportunities to learn and attain the skills they require for their chosen career path, and to provide a welcoming and safe place to excel.
If you are interested in a postdoctoral position, unless otherwise posted, it is typically required to apply to external funding. There are multiples sources available from the University (including the Data Sciences Institute, and the Faculty of Arts & Science, among others) and the Federal and Provincial Governments in Canada (eg NSERC and CIHR). Funding is also available from some international agencies (e.g. HFSP, Marie-Curie), and for citizens of specific countries (USA, China, etc). If you join my lab as a Postdoctoral fellow, my philosophy is to guide and help you design a competitive and fund-able research program, therefore, I am happy to work with soon-to-be PhD graduates to design a project and apply for funding.
Graduate students can currently apply through the Cell & Systems Biology graduate program at either the MSc or PhD levels. Please reach out to Max as early as possible in the Fall or Winter prior to when you want to join the lab to discuss opportunities and available projects. If you are an international applicant, please be aware that there are limited and very competitive spots in the program for non-Canadian residents (or permanent residents).
I am typically interested in hosting 1-3 undergraduate students per academic cycle (depending on availability of projects). These can be in the form of the Research Opportunities Program, workstudy positions, through an independent project in CSB (CSB497/498) or BCB (BCB330/430), or through the NSERC USRA program / CSB Summer studentship. Interested students should reach out Max as early as possible to discuss interests and the potential availability of projects. More information on undergraduate research opportunities can be found on the Cell & Systems Biology website.

